Fmoc Solid Phase Peptide Synthesis
Contents
1. The Fmoc Group
1.1. General Remarks
1.2. Cleavage Procedures
2. Coupling Reaction
2.1. General Remarks
2.2. Activation Methods
2.2.1. Carbodiimide-Carbodiimide/HOBt
2.2.2. Activation by Phosphonium and Uronium/Aminium Salts
2.2.3. Fmoc Amino Acid Active Esters
2.2.4. Fmoc Amino Acid Chlorides and Fluorides
2.3. Monitoring of Coupling and Deblocking
2.3.1. The Kaiser Test
2.3.2. The TNBS Test
2.3.3. The Acetaldehyde/Chloranil Test
2.3.4. The Bromophenol Blue Test
2.3.5. Cleavage of Samples
2.4. Capping
2.5. Aggregation/Difficult Sequences
3. Cleavage from the Resin
3.1. Simultaneous Cleavage from the Resin/Side-Chain Deprotection
3.2. Mix Your Own Cocktail
3.3. Cleavage of Protected Peptide Fragments
4. Side Reactions in Fmoc SPPS
4.1. Diketopiperazine Formation
4.2. Aspartimide Formation
4.3. Transfer of Pmc from Arg to Trp During TFA Deprotection
4.4. 3-(1-Piperidinyl)alanine Formation
4.5. Incomplete Fmoc Deprotection
4.6. Guanylation
5. References
6. Related Products
7. Useful Links
1.1. General Remarks
Due to the development of strategies based on orthogonal protection, Fmoc has become the most important base-labile Nα-protecting group.
Fmoc is acid-stable, withstands cleavage of Boc/tBu (TFA) and Z/Bzl (HF). Fmoc is stable under the cleavage conditions of Aloc/All (Pd°). Towards tertiary amines such as DIPEA, pyridine [1]; the relative stability depends on base concentration, solvent and temperature. Stability towards hydrogenolysis is controversial [2] and should be evaluated for each individual case. Towards bases, especially secondary amines [1] (piperidine>diethylamine); fortunately the Fmoc group is less labile towards primary amines, including the amino group of the amino acid involved in the coupling reaction; premature Fmoc cleavage may nevertheless occur during very slow couplings. N-silylation of the coupling site prevents this side reaction and can enhance the coupling [3].
The Fmoc group is removed via base-induced β-elimination. As a result dibenzofulvene and carbon dioxide are split off. Secondary bases such as piperidine add to the former molecule whereas bases such as DBU do not react with the dibenzofulvene which has to be removed rapidly from the peptide resin or scavenged by a secondary amine such as piperidine to avoid irreversible attachment to the liberated amino group. Since both cleavage products are strong chromophors the deblocking can be monitored by UV spectroscopy.
By-products generated by the repetitive treatment with base have been described. Aspartimide formation is the best documented side-reaction. Epimerization and subsequent piperidide formation have been detected, even though the bulky tert. butyl group impedes reactions involving the β-carboxy group. Aggregation during chain elongation interferes with the couplings as well as with the Fmoc cleavages. If an incomplete deblocking occurs or is suspected, more active cleavage reagents should be tested.
1.2. Cleavage Procedures
Usually, Fmoc is split off by a short treatment (3 to 5 minutes) with piperidine/DMF (1:4). In general this treatment is repeated and slightly prolonged (7 to 10 minutes). Under those conditions complete deblocking is attained in most cases. Thus, deviations are restricted to cases of sluggish cleavage or base-sensitive sequences.
Harsher alternatives to piperidine/DMF have been developed as well as milder cleavage reagents [4]. In case of sluggish deblocking, even slight variations of the reagent may considerably accelerate the cleavage, e.g.:
– 1 to 5% DBU/DMF, more reactive than piperidine [5], for glycopeptides [6],
– 20% piperidine and 1-5% DBU in DMF, for difficult deblockings,
– morpholine/DMF (1:1), milder than piperidine for highly sensitive glycopeptides [7],
– piperidine/DMF (1:4) at 45°C, for “difficult sequences” [8],
– 0.1 M HOBt in piperidine/DMF (1:4), suppression of DKP and aspartimide formation [9],
– Bu4N+F- in DMF and other tetraalkylammonium fluorides [10] (not recommended),
– 2% HOBt, 2% hexamethyleneimine, 25% N-methylpyrrolidine in DMSO/NMP 1:1, mild cleavage keeping thioesters intact [11].
But, whichever cleavage reagent is preferred, it has to be washed out very carefully after Fmoc removal, the last washing must be neutral. When synthesizing large peptides the duration of Fmoc cleavage should be gradually increased. For safe removal of the deblocking reagent the resin may have to be washed more often.
RECOMMENDED STANDARD PROCEDURE
Fmoc Cleavage
Prewash with DMF (2x)
|
As mentioned above, the generation and disappearance of Fmoc based chromophors allows the monitoring of the synthesis. Furthermore, samples may be taken to determine the load of Fmoc peptide. The completion of the deprotection reaction may be checked by cleaving samples and analyzing the obtained peptide.
RECOMMENDED STANDARD PROCEDURE
Determination of Load
A sample of peptide-resin is washed 4xDMF, 5xIPA and 2xMeOH or ether,
|
RECOMMENDED STANDARD PROCEDURE
Determination of the Completion of Fmoc Cleavage
The sample is washed and dried as described above.
|
2. Coupling Reaction
2.1. General Remarks
When considering the general coupling reaction in an SPPS process, its efficacy is limited by a set of variables such as:
– the choice of the solvent. It will in part determine the swelling of the peptide-resin and influence the accessibility to the reactive sites; it will also have a direct effect on the kinetics of the coupling reaction.
– the steric hindrance. It is determined by the nature of the side chains R1 and R2 and of their protecting groups.
– the reactivity of the activated carboxylic acid.

It is an important variable because it will allow the rapid acylation of the amino function and have a limiting effect on possible side reactions.
Generally speaking, the first priority is to reach the optimal combination of high coupling rate and minimal racemization. Indeed contrary to widespread assumptions Nα-urethane protected amino acids can racemize considerably under inadequate coupling conditions [12,13,14].
“Standard” coupling conditions may result in incomplete coupling and formation of by-products such as epimers, truncated peptides and sequences, products of guanylation [15], or products generated by premature Fmoc cleavage. A more potent coupling reagent such as HATU [16] or very active Fmoc amino acid denyatives such as the acid fluorides [17] may drive the coupling to completion, N-silylation with bis(trimethylsilyl)acetamide (the use of TMS-Cl is restricted to Boc-SPPS) [3,18] may be an additional means to promote the reaction.
Better results will be obtained by repeating a coupling with fresh reagents (and changing coupling parameters if a low conversion was obtained) rather than by prolonging the reaction. Generally, coupling protocols may be changed in the course of a synthesis, especially when optimizing an SPPS.
A special paragraph will be dedicated to the problems caused by peptide aggregation in the course of the synthesis. This phenomenon is a major cause of trouble as it is difficult to predict, is sequence dependent and no universal solution has been found up to now.
Numerous ways are available for the activation of the carboxylic function and we want to limit our discussion to the most widely used ones in SPPS.
2.2. Activation Methods
2.2.1. Carbodiimide – Carbodiimide/HOBt
DCC activation has been used from the first days of the solid phase technique [19] and is still popular today. DIC [20] is also frequently used and presents the advantage that the corresponding urea is more soluble than the one obtained from DCC.
However the activation by carbodiimides presents several drawbacks:
The coupling (acylation of the amino function) is not particularly rapid and the activated species can be deactivated through rearrange into an N-acyl urea;

Racemization can occur following cyclization of the activated species to a 5(4H)-oxa-zolone which racemizes via enolization and the subsequent reopening by the amino component yields epimers.

These problems can usually be avoided by the introduction of additives to the reaction mixture. The most popular additive in SPPS is HOBt [21].
In a standard coupling procedure the HOBt ester is generated by the reaction between the protected amino acid and HOBt. The reaction is mediated by DCC or DIC.
Recently HOAt, an aza analogue of HOBt has been reported to markedly accelerate the acylation and also to reduce racemization [22,23]. This new additive is widely expected to overcome problems related to “difficult” coupling.
Whereas DCM proves to be the optimal solvent for carbodiimide activation it is less suitable for the subsequent coupling step which proceeds more rapidly in more polar solvents such as DMF. Therefore a mixture of DCM and DMF is often used for both the activation and the coupling steps.
RECOMMENDED STANDARD PROCEDURE
Activation and Coupling with DCC/HOBt
After deprotection of the amino function the resin is thoroughly washed.
|
CAUTION: DCC is an aggressive allergen. Inhalation and contact must be avoided; adequate protection must be worn when working with DCC and efficient ventilation is also required!
The preactivation time must be kept at a minimum when generating the Fmoc-Arg(Pmc,Pbf)-OBt derivative as the activated Arg derivatives may cyclize yielding an unreactive lactam.
The same is true for the activation of Fmoc-His(Trt)-OH since racemization catalyzed by the nitrogen of the imidazole ring may occur. For the coupling of especially bulky amino acids such as Aib, Tic, … or in the case of recognized difficult coupling we recommend the replacement of HOBt by HOAt, or the use of other activating agents.
2.2.2. Activation by Phosphonium and Uronium/Aminium Salts
In recent years acylphosphonium (BOP, PyBOP) [24,25] and acyluronium/aminium salts (HBTU, TBTU) [26] have become extremely popular coupling agents in SPPS.
TBTU converts the Fmoc amino acid into the active OBt ester in the presence of one equivalent of base. DIPEA is routinely used but it has recently been demonstrated that it can induce racemization in the coupling of Fmoc-Cys(Trt)-OH [13] and of Fmoc-Ser(tBu)-OH [14]. The use of collidine has been recommended as a substitute for DIPEA [27]. Nevertheless large excess of base is to be avoided even though coupling is accelerated at a slightly basic pH.
For the coupling of very bulky amino acids such as N-alkylamino acids or α-dialkylamino acids, the moderately reactive TBTU should be replaced by more potent reagents such as HATU [16], TATU, or PyBOP [25].
RECOMMENDED STANDARD PROCEDURE
TBTU/DIPEA (collidine) Activation
The resin is prepared as described in the previous procedure.
|
The comments concerning the preactivation of Fmoc-Arg (Pmc, Pbf)-OH and FmocHis(Trt)-OH are also valid in this case.
It is necessary to carry out the preactivation step when working with uronium derivatives such as TBTU or TATU as they can react with the free amino group of the peptide-resin to yield substituted guanidines [15].
2.2.3. Fmoc Amino Acid Active Esters
Active esters have been used well before the introduction of the solid phase technique. Several classes of active esters are sufficiently stable for isolation, prolonged storage and still react rapidly in peptide coupling. Among them Fmoc-AA-OPfp esters [28] and Fmoc-AA-ODhbt esters [29] have become popular in automated SPPS, they are merely dissolved and added to the resin (in case of Pfp esters the addition of HOBt to increase their reactivity has been recommended [30]).
Also small scale manual SPPS as well as multiple peptide synthesis and the synthesis of pep- tide libraries can be performed very rapidly and conveniently with preformed active esters. Fmoc-AA-ONp and Fmoc-AA-OSu have found only restricted application in SPPS.
2.2.4. Fmoc Amino Acid Chlorides and Fluorides
The crystalline, rather stable Fmoc amino acid fluorides have recently gained much interest [94]. They may also be obtained in situ by treating the Fmoc amino acids with TFFH [110]. Even Fmoc amino acids bearing acid-labile side-chain protection can be converted into stable acid fluorides (except for Arg and His derivatives for which the corresponding acid fluorides should only be generated in situ). The in situ preparation of the Fmoc amino acid fluorides is made by reaction with TFFH [31] or BTFFH [32].
Fmoc amino acid chlorides can also be used but their applicability is more limited [33]; also acid-sensitive groups may not be present when treating Fmoc amino acids with oxalyl chloride or thionyl chloride to obtain the acid chlorides [34,35].
A recent publication has described the in situ preparation of Fmoc amino acid chlorides by reaction with bis(trichloromethyl) carbonate and their use in difficult couplings [36].
2.3. Monitoring of Coupling and Deblocking
In Fmoc SPPS the monitoring of the completion of the Fmoc cleavage and of the coupling reaction is essential.
Reliable methods detecting minute amounts of unreacted amino groups are essential for the monitoring of the coupling reaction. On the other hand the ability of detecting small quantities of Fmoc peptide allows the control of the completion of the Fmoc cleavage.
A range of color tests for the qualitative monitoring of the coupling reaction has been developed and we describe the most commonly used ones here below. Each one of these tests having its limitations, we recommend to perform two independent tests in the monitoring of the completion of the coupling.
It must also be noted that during the synthesis of very long peptides the detection of the remaining free amino group will usually become difficult with increasing peptide length.
The same is true when aggregation phenomenon occurs, limiting the accessibility of the amino function.
In these cases it is recommended to use a protocol including systematic double coupling followed by a capping step.
The tests described here below can then be performed with small samples of washed resin.
It is recommended to perform control tests with protected amino function (negative control) and deprotected amino function (positive control).
2.3.1. The Kaiser Test
The Kaiser test [37] is based on the reaction of ninhydrin with amines. It is a very sensitive test for primary amines, visualized by an intense blue color, and somewhat less suitable for secondary amines, being detected by development of a brownish red color.
Usually the color is developed mainly in the beads and partly in the supernatant; but for spectrometric quantitative determination of the amount of unreacted amino groups the color has to be transferred completely to the solution [38]. The intensity of the color depends on the nature of the amino terminus to be detected; rather unspecific shades are obtained with N-terminal (side-chain protected!) Asp, Asn, Cys, Ser, and Thr. Brownish red beads result with N-terminal Pro. As the resin sample has to be heated, “hidden” NH2-groups may become more accessible and thus detectable. However, prolonged heating as well as overheating should be avoided as it may cause Lys(Boc) cleavage or Fmoc removal (by pyridine).
RECOMMENDED STANDARD PROCEDURE
Kaiser Test
Solution 1: 5 g ninhydrin in 100 ml ethanol
|
The Kaiser test can also be used for the quantitative monitoring of the coupling [38] even if quantitation is not indispensable in routine SPPS.
2.3.2. The TNBS Test
The TNBS test [39] is easier to perform and of a nearly equivalent sensitivity as the Kaiser test. It can only be used for detecting primary amino groups. Only the beads will turn orange-red and the intensity of the color does not depend on the nature of the N-terminal amino acid. In the monitoring of the completion of the coupling, it is recommended to inspect the beads under a microscope. Indeed, light orange color in the core of the beads cannot be detected by visual inspection.
We recommend the use of the combination Kaiser/TNBS tests in the routine detection of primary amino function.
RECOMMENDED STANDARD PROCEDURE
TNBS test
Solution 1: 10% DIPEA in DMF
|
2.3.3. The Acetaldehyde/Chloranil Test
This sensitive test has been developed for reliable detection of secondary amino groups [40] but it will also detect primary amines. As for the TNBS test, only the beads will be colored in case of a positive test, they should be inspected under a microscope.
RECOMMENDED STANDARD PROCEDURE
Acetaldehyde/Chloranil Test
The stock solutions should be kept in the refrigerator and for one month maximum.
|
2.3.4. The Bromophenol Blue Test
As all types of amines can be detected applying this test [41] traces of DIPEA or collidine have to be removed very carefully.
Traces of acid lead to false negative results. The beads may be inspected under a microscope.
Minute amounts of bromophenol blue have been added to couplings in progress to monitor the conversion; the end-point is indicated by discoloration [42].
RECOMMENDED STANDARD PROCEDURE
Bromophenol Blue Test
Reagent solution: 0.05% bromophenol blue in DMA
|
2.3.5. Cleavage of Samples
Cleavage of samples may seem a rather elaborate way of monitoring an SPPS, but it is the most comprehensive method. It is especially useful when the synthesis has to be optimized or documented or when negative color tests results are obtained because of scarcely accessible coupling sites.
The cleavage of a sample is also a reliable method to monitor the completion of the Fmoc cleavage.
After the cleavage the sample is analyzed by HPLC as Fmoc protected and free amino sequences are usually well separated in a standard HPLC gradient.
If necessary, additional information can be gained from mass spectrometry or microsequencing.
This approach can even be further simplified and accelerated, as MALDI-MS turned out to be a simple and very effective method for monitoring an SPPS as only a few beads of peptide resin have to be cleaved to perform a mass spectrum [43,44,45].
RECOMMENDED STANDARD PROCEDURE
TFA Cleavage of In Process Samples
We recommend the use of a standard cleavage cocktail
|
2.4. Capping
If the color tests are positive after the predetermined coupling time the resin is filtered off, washed with DMF and IPA and a second coupling is usually performed.
If the test is slightly positive the recoupling is usually made using a smaller amount of reagents, typically 50% of the quantities used in the first coupling.
One or two parameters may be changed in the second coupling however changes of coupling conditions are certainly due if positive tests are obtained after the second coupling!
Some possible changes
– Change solvent: DMF/DCM 1:1 instead of DMF,
– Change coupling reagent: TATU instead of TBTU,
– Change additive: TBTU+HOAt instead of TBTU+HOBt.
When the color tests keep revealing the presence of unreacted amino functions after recoupling, it is necessary to cap these to avoid the formation of deletion sequences. The capping will yield a truncated sequence (shortened pep- tide) but the truncated sequences differ generally considerably from the final peptide and can be readily separated.
Capping is realized through a short treatment of the peptide resin with a large excess of a highly reactive unhindered acid derivative and abase, usually acetic anhydride or benzoyl chloride and pyridine. At the end of the capping step the reagents are filtered off and the resin is carefully washed before proceeding to the next deprotection step.
For the synthesis of long peptides we recommend to use a protocol in which systematic double coupling is followed by capping.
RECOMMENDED STANDARD PROCEDURE
Capping
After filtration of the coupling solution the peptide-resin is washed several times with DMF, IPA;
|
Capping can also be used to decrease the load of a resin.
For that purpose a deficient amount of the C-terminal or penultimate Fmoc amino acid is coupled to the unloaded or preloaded resin. The resulting load is determined and when the desired level of substitution has been reached the remaining free amino groups are blocked by acetylation.
2.5. Aggregation/Difficult Sequences
Repeated incomplete deprotection of the αamino function as well as difficulties in obtaining a complete coupling reaction are some of the problems caused by the on resin aggregation of the peptide chain.
This phenomenon has been attributed to the self-association of the peptide chain by formation of hydrogen bonds [46].
It is sequence dependent and up to now no “Golden Rule” exists which allows the prediction of aggregation during the synthesis of a new sequence!
Some leads have however been listed but they can just be looked at as informative. The aggregation has a tendency to occur in a sequence of hydrophobic residues; it has been described as starting from the 5/6th residue from the C-terminus but not occurring after the 21st one [47].
Two approaches can be used to disrupt the aggregation; the first one consists in modifying the environment in which the synthesis is made and to introduce elements known to disrupt hydrogen bonds; among them we list:
– Addition of solvent such as DMSO [48],
– Addition of chaotropic salts [49],
– Addition of nonionic detergents and of ethylene carbonate (“Magic Mixture”) [50],
– Coupling at elevated temperature [51],
– Sonication of the coupling reaction [52],
– Reduction of the peptide load on the resin,
The other approach is to introduce backbone protecting groups which will prevent the formation of hydrogen bonds. Such protection is made by the introduction of the Hmb group on the αnitrogen [53]. It has been shown that the presence of a Hmb unit every 6-7 residues is sufficient to disrupt the peptide aggregation [54]. The Hmb protected amino acid is introduced under the form of N,O-bis-Fmoc-N-(2hydroxy-4-methoxybenzyl) derivative, the O-Fmoc protection being cleaved during the following piperidine treatment. At the end of the synthesis the Hmb group is cleaved in the final TFA cleavage.
One other possibility of protection of the αnitrogen is to introduce dipeptides possessing a pseudoproline residue, derived from Set or Thr since Pro is known to disrupt aggregation [55].
At the end of the synthesis the Ser or Thr will be regenerated in the final TFA cleavage step.
3. Cleavage from the Resin
3.1. Simultaneous Cleavage from the Resin/Side-Chain Deprotection
Concentrated TFA (95% aqueous TFA) is the typical reagent to perform the final cleavage of the peptide from the resin together with the removal of the side chain protecting groups.
During the reaction highly reactive carbocations are generated and it is necessary to trap them to avoid undesired reactions with sensitive amino acids such as Cys, Met, Set, Thr, Trp, Tyr.
This effect is obtained by the addition of scavengers to the cleavage solution.
Water is a moderately efficient scavenger and can be used as single scavenger for the cleavage of peptides devoid of Cys, Met and Trp.
EDT and DTE are the most common and efficient scavengers for peptides containing sensitive amino acids.
Several papers have been published analyzing the advantages of different cleavage cocktails [56,57] and recommending:
Reagent K: TFA/thioanisole/water/phenol/EDT (82.5:5:5:5:2.5 v/v)
The use of sulfur containing scavengers is recommended in the cleavage of sequences containing Met, Cys and Arg. Care must be taken in the use of thioanisole as it has been suggested that it participates in the premature cleavage of Acm, StBu, or tBu from Cys residues [58].
Also silane derivatives (TES, TIS) can successfully replace the malodorous EDT [59,60,61]. They show good efficacy in quenching carbocations in sequences containing Arg and Trp residues.
It is strongly advised to perform sample cleavages using different cocktail compositions and to compare the qualitative and quantitative recovery before choosing the final one. The monitoring of these sample cleavage is best done by analytical HPLC.
In the development of the cleavage conditions it is also advisable to perform a simultaneous time course evaluation of the evolution of the cleavage. It should be complete in 2-3 hours at room temperature in order to minimize side reactions arising from long exposure to strong acid environment.
The table below shows the recommended scavengers in relation to the amino acids present in the sequence.
3.2. Mix Your Own Cocktail
Sample and bulk cleavages should be performed in a well ventilated hood and protective equipment (gloves, goggles, mask if necessary) worn by the personnel performing the operation.
Starting Cocktail: TFA/water 95:5
Add:
|
Amino acids present |
Recommended scavengers |
|
Cys |
DTE, EDT |
|
Met |
DTE, EDT, 2-mercaptoethanol, ethyl methyl sulfide |
|
Ser, Thr, Tyr |
m- or p-cresol, DTE, EDT |
|
Trp (unprotected) |
DTE, EDT, 2-Me-indole, Ac-Trp-OMe, tryptamine, TIS |
A growing proportion of scavengers means reduction of the amount of TFA, thus, cleavage with cocktails containing a considerable amount of scavenger or many different scavengers may take longer.
Having optimized the composition of the cocktail and the reaction time it is possible to perform the cleavage of the bulk of peptide resin.
One of the advantages of the Fmoc SPPS is the possibility of scaling up the cleavage batch size without special equipment.
RECOMMENDED STANDARD PROCEDURE
TFA Cleavage
The cleavage cocktail (10-15 mL/g peptide-resin) is prepared in a reactor or a
|
Filtrates containing sulfur scavengers have to be oxidized to eliminate the pervasive obnoxious smell. For that purpose the TFA filtrate is neutralized (NaOH/ice) before addition of an oxidant (e.g. bleach or a saturated solution of iodine in ethanol).
3.3. Cleavage of Protected Peptide Fragments
A range of cleavage reagents for peptides synthesized on 2-chlorotrityl resin has been described. TFE/AcOH/DCM (1:1:3) has been developed by Barbs [62]. Cleavage is also rapidly attained with 0.5% TFA/DCM as well as with HFIP/DCM (1:4 or 3:7) [63].
The latter system is especially suitable for the cleavage of fully protected fragments to be coupled to another fragment in solution, as it eliminates the use of a carboxylic acid in the cleavage step.
RECOMMENDED STANDARD PROCEDURE
Cleavage from 2-Chlorotrityl Resin
The peptide-resin is stirred for 45-60 minutes in HFIP/DCM 1:4 (10 mL/g peptide-resin).
|
4. Side Reactions in Fmoc SPPS
4.1. Diketopiperazine Formation
In peptide synthesis diketopiperazine formation is a notorious side-reaction at the dipeptide stage and is particularly prone to occur in Fmoc based SPPS because of its mechanism.
During the base-induced deprotection of the penultimate amino acid, the liberated amino function may cleave the resin ester linkage whilst forming a piperazinedione, i.e. the dipeptide is split off.
Good accessibility to the ester bond facilitates this side-reaction as does a cis-amide bond making sequences containing a C-terminal Pro prone to this side-reaction. Nearly quantitative loss may occur.

The extent of diketopiperazine formation depends on the nature of the C-terminus as well as on the penultimate amino acid, especially on the bulk of the side-chains.
Diketopiperazine formation may be circumvented in Fmoc/tBu SPPS
– by using 2-chlorotrityl chloride resin or other bulky resins such as DHPP-Resin.
– by coupling the appropriate Fmoc-dipeptide in lieu of the penultimate amino acid. In that case a risk of epimerization exists if the C-terminal amino acid of this dipeptide is not Gly or Pro.
4.2. Aspartimide Formation
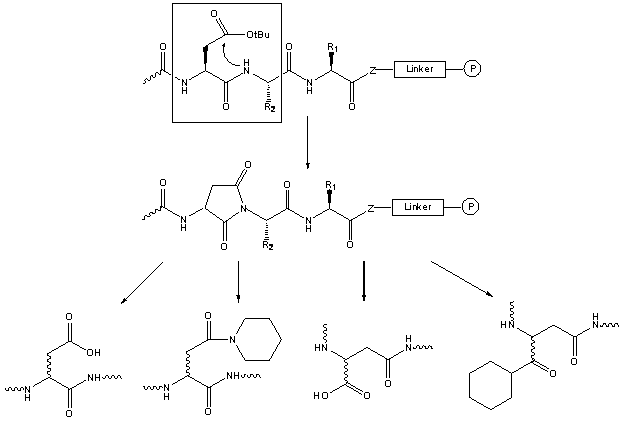
This sequence-dependent side-reaction is catalyzed by acids as well as by bases.
In Fmoc based SPPS aspartimide can thus be generated during the final TFA cleavage as well as during the chain elongation; this cyclization is accompanied by epimerization.
The tert.butyl side-chain protecting group normally provides sufficient protection.
Several examples of aspartimide formation during the peptide elongation have been reported, together with the reopening of the aspartimide cycle during the subsequent Fmoc deprotection step, yielding piperidides [64,65,66].
The addition of HOBt to the piperidine solution helps to reduce the formation of aspartimide. Another approach is to introduce the residue preceeding Asp as the Fmoc(Hmb) protected amino acid [67].
4.3. Transfer of Pmc from Arg to Trp During TFA Deprotection
This reaction occurs when Trp is used unprotected. The addition of Pmc (or Pbf) to the indole ring will occur and the amount of by-product is related to the distance between the Trp and the Arg residues [68].
4.4. 3-(1-Piperidinyl)alanine Formation
This modified amino acid is obtained when synthesizing peptides with C-terminal Cys.
The base catalyzed elimination of the suffhydryl protecting group affords dehydroalanine and the subsequent addition of piperidine yields the C-terminally modified peptide [69]. This side reaction is minimized (but not avoided!) when trityl is used for the protection of the C-terminal Cys.

4.5. Incomplete Fmoc Deprotection
It has been demonstrated [2] that the incomplete deprotection is associated with slow or incomplete coupling. In such cases, prolonged or repeated piperidine treatment, eventually associated with a change in solvent can help to reduce the problem.
Again this phenomenon is difficult to predict. Therefore the necessity of careful monitoring of the completion of the cleavage when doubts arise is mandatory.
4.6. Guanylation

This reaction occurs during a coupling induced by uronium/arniniurn reagents. This side reaction is avoided when the incoming amino acid is preactivated.
5. References
[1] E. Atherton, RC. Sheppard in The Peptides vol. 9, S. Udenfriend, J. Meienhofer Eds, Academic Press, New York 1987, 1.
[2] E. Atherton, C. Bury, R.C. Sheppard, Bj. Williams, Tetrahedron Lett. (1979) 3041.
[3] H. Wenschuh, M. Beyermann, R Winter, M. Bienert, D. lonescu, L.A. Carpino, Tetrahedron Lett. 37 (1996) 5483.
[4] G.B. Fields in Peptide Synthesis Protocols, M.W. Pennington, B.M. Dunn Eds, Humana Press, Totowa NJ 1994, 17.
[5] J.D. Wade,J. Bedford, R.C. Sheppard, G.W. Tregear, Pept. Res. 4 (1991) 194.
[6] M. Meldal, T. Bielfeldt, S. Peters, Kj. Jensen, H. Paulsen, K. Bock, mt. J. Pept. Protein Res. 43 (1994) 529.
[7] B. Liebe, H. Kunz,Angew. Chem. Tnt. Ed. Engl. 36 (1997) 618.
[8] B.D. Larsen, A. Holm, Int.J. Pept. Protein Res. 43 (1994) 1.
[9] R. Dolling, M. Beyermann,J. Haenel, F. Kernchen, E. Krause, P. Franke, M. Brudel, M. Bienert,J. Chem. Soc., Chem. Comm. (1994) 853.
[10] A. Kapurniotu, C. Ungermann, W. Voelter, in 2nd International Symposium on Innovation and Perspectives in SPPS, Canterbury 1991, R. Epton Ed., Intercept Ltd, Andover 1992, 319.
[11] X. Li, T. Kawakami, S. Aimoto, Tetrahedron Lett. 39 (1998) 8669.
[12] N.L. Benoiton, K. Kuroda, Int.J. Pept. Protein Res. 17(1981)197.
[13] Y. Han, F. Albericio, G. Barany,J. Org. Chem. 62 (1997) 4307.
[14] A. di Fenza, M. Tancredi, C. Galoppini, P. Rovero, Tetrahedron Lett. 39 (1998) 8529.
[15] F. Albericio,J.M. Bofill, A. El-Faham, S.A. Kates,J. Org. Chem. 63 (1998) 9678.
[16] L.A. Carpino,A. El-Faham,J. Org. Chem. 59 (1994) 695.
[17] H. Wenschuh, M. Beyermann, E. Krause, M. Brudel, R. Winter, M. Schumann, L.A. Carpino, M. Bienert,J. Org. Chem. 59 (1994) 3275.
[18] A. Brunissen, M. Ayoub, S. Lavielle, Tetrahedron Lett. 37 (1996) 6713.
[19] RB. MerrifIeld, Biochemistry 3 (1964) 1385.
[20] D. Sarantakis,J. Teichman, El. Lien, R Fenichel, Biochem. Biophys. Res. Comm. 73 (1976) 336.
[21] W. Konig, R. Geiger, Chem. Ber. 103 (1970) 788.
[22] L.A. Carpino,J. Am. Chem. Soc. 115 (1993) 4397.
[23] LA. Carpino, A. El-Faham, C.A. Minor, F. Albericio,J. Chem. Soc., Chem. Comm. (1994) 201.
[24] B. Castro,J.-R Dormoy, G. Evin, C. Selve, Tetrahedron Lett. (1975) 1219.
[25] J. Coste, D. Le-Nguyen, B. Castro, Tetrahedron Lett. 31 (1990) 205.
[26] R Knorr, A. Trzeciak, W. Bannwarth, D. Gillessen, Tetrahedron Lett. 30 (1989) 1927.
[27] L.A. Carpino, D. lonescu, A. El-Faham,J. Org. Chem. 61 (1996) 2460.
[28] L. Kisfaludy, I. Schön, Synthesis (1983) 325.
[29] E. Atherton,J.L. Holder, M. Meldal, R.C. Sheppard, RM. Valerio,J. Chem. Soc., Perkin Trans. 1 (1988) 2887.
[30] E. Atherton, L.R Cameron, R C. Sheppard, Tetrahedron 44 (1988) 843.
[31] L.A. Carpino, A. El-Faham,J. Am. Chem. Soc. 117(1995) 5401.
[32] A. El-Faham, Chemistry Letters (1998) 671.
[33] L.A. Carpino, M. Beyermann, H. Wenschuh, M. Bienert,Acc. Chem. Res. 29 (1996) 268.
[34] L.A. Carpino, Bj. Cohen, K.E. StephensJr, S.Y. Sadat-Aalaee,J.H. Tien, D.C. Langridge, J. Org. Chem. 51 (1986) 3732.
[35] M. Beyermann, M. Bienert, H. Niedrich, L.A. Carpino, D. Sadat-Aalaee,J. Org. Chem. 55 (1990) 721.
[36] E. FaIb, T. Yechezkel, Y. Salitra, C. Gilon,J. Peptide Res. 53 (1999) 507.
[37] E. Kaiser, RL. Colescott, C.D. Bossiriger, P.1. Cook, Anal. Biochem. 34 (1970) 595.
[38] V.K. Sarin, S.B.H. Kent,J.P. Tarn, RB. Merrifleld, Anal. Biochem. 117(1981) 147.
[39] W.S. Hancock,J.E. Battersby, Anal. Biochem. 71 (1976) 260.
[40] T. Vojkovsky, Pept. Res. 8 (1995) 236.
[41] V. Krchnãk,J. Vágner, P. àfár, M. Lebi, Coil. Czech. Chern. Comm. 53 (1988) 2542.
[42] V. Krchnák,J. Vágner, M. LebI, Int.J. Pept. Protein Res. 32 (1988) 415.
[43] B.J. Egner, Gj. Langley, M. Bradley,J. Org. Chem. 60(1995)2652.
[44] G. Talbo,J.D. Wade, N. Dawson, M. Manoussios, G.W. Tregear, Lett. Pept. Sci. 4(1997)121.
[45] C.L. Brummel,J.C. Vickerman, S.A. Carr, M.E. Hemling, G.D. Roberts, W.Johnson, J. Weinstock, D. Gaitanopoulos, Sj. Benkovic, N. Winograd, Anal. Chern. 68 (1996) 237.
[46] E. Giralt,J. Rizo, E. Pedroso, Tetrahedron 40 (1984) 4141.
[47] S.M. Meister, S.B.H. Kent, in Peptides, Chemistry Structure and Biology, Proceedings 8th American Peptide Symposium Tucson, 1983, Vj. Hruby, D.H. Rich Eds, Pierce Chemical Company, Rockford Il 1983, 103.
[48] E. Oliveira, A. Miranda, F. Albericio, D. Andreu, A.C.M. Paiva, C.R. Nakaie, M. Torninaga, J. Pept. Res. 49 (1997) 300.
[49] M.W. Pennington, I. Zaydenberg, M.E. Byrnes, RS. Norton, W.R Kern, Tnt. J. Pept. Protein Res. 43 (1994) 463.
[50] L. Zhang, C. Goldammer, B. Henkel, F. Zühl, G. Panhaus, G.Jung, E. Bayer, in 3rdlnternational Symposium on Innovation and Perspectives in SPPS, Oxford UK, R Epton Ed., Mayflower Scientific Ltd, Birmingham 1994, 711.
[51] L.M. Varanda, M.T.M. Miranda,J. Pept. Res. 50 (1997) 102.
[52] H.G. Chao, M.S. Bernatowicz, G.R. Matsueda,J. Org. Chem. 58 (1993) 2640.
[53] M. Quibell, Tjohnson, in Fmoc Solid Phase Peptide Synthesis-A Practical Approach, W.C. Chan, P.D. White Eds, Oxford University Press 2000, 115, and references therein.
[54] C. Hyde, Tj.Johnson, D. Owen, M. Quibell, R.C. Sheppard, Int.J. Pept. Protein Res. 43 (1994) 431.
[55] M. Mutter, A. Nefzi, T. Sato, X. Sun, F. Wahl,T. Wohr, Pept. Res. 8 (1995) 145.
[56] D.S. King, C.G. Fields, G.B. Fields, Tnt.J. Pept. Protein Res. 36 (1989) 255.
[57] F. Dick in Peptide Synthesis Protocols, M. Pennington and B.M. Dunn Eds, Humana Press, Totowa NJ 1994, 63.
[58] E. Atherton, R.C. Sheppard, P. Ward,J. Chem. Soc., Perkin Trans. I (1985) 2065.
[59] D. Pearson, M. Blanchette, M.L. Baker, C.A. Guidon, Tetrahedron Lett. 30 (1989) 2739.
[60] C.G. Fields, G.B. Fields, Tetrahedron Lett. 34 (1993) 6661.
[61] N.A. Sole, G. Barany,J. Org. Chem. 57(1992) 5399.
[62] K. Barbs, D. Gatos, J.Kallitsis, G. Papaphotiu, P. Sotiriu, Y. Wenging, W. Schafer, Tetrahedron Lett. 30(1989) 3943.
[63] R Bolihagen, M. Schmiedberger, K. Barbs, E. Grell,J. Chem. Soc., Chem. Comm. (1994) 2559.
[64] R Dolling, M. Beyermann,J. Haenel, F. Kernchen, E. Krause, P. Franke, M. Brudel,
M. Bienert in 3rd International Symposium on Innovation and Perspectives in SPPS, Oxford UK, R. Epton Ed., Mayflower Scientific Ltd, Birmingham 1994,489.
[65] Y. Yang, W.V. Sweeney, K. Schneider, S. Thornqvist, B.T. Chait, J.P. Tam, Tetrahedron Lett. 52 (1994) 9689.
[66] J.L. Lauer, C.G. Fields, G.B. Fields, Lett. Pept. Sci. 1 (1994) 197.
[67] M. Quibell, D. Owen, L.C. Packman, Tj.Johnson,J. Chem. Soc., Chem. Comm. (1994) 2343.
[68] A. Stierandova, N.F. Sepetov, G.V. Nikiforovich, M. LebI, Int.J. Pept. Protein Res. 43 (1994) 31.
[69] J. Lukszo, D. Patterson, F. Albericio, S.A. Kates, Lett. Pept. Sci. 3 (1996) 157.
7. Useful Links
Peptide Synthesis
Introduction to Peptide Synthesis
Fmoc Solid Phase Peptide Synthesis
Fmoc Solid Phase Peptide Synthesis: A Practical Approach
Methods and protocols of modern solid phase Peptide synthesis
Linkers, resins, and general procedures for solid-phase peptide synthesis
Handles for Fmoc solid-phase synthesis of protected peptides
Peptides, solid-phase synthesis and characterization: Tailor-made methodologies
Solid-phase peptide synthesis: from standard procedures to the synthesis of difficult sequences


