Overview of Antibody
This entry is from Wikipedia, the leading user-contributed encyclopedia.
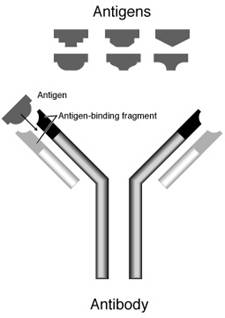
An antibody or immunoglobulin is a large Y-shaped protein used by the immune system to identify and neutralize foreign objects like bacteria and viruses. The antibody recognizes a specific target, which is called an antigen.[1] An antibody contains two sites called paratopes that bind antigens. These structures can be thought of as similar to locks and are specific for just one particular part of the antigen called an epitope, which can be thought of as similar to a key. This specific lock and key interaction allows an antibody to tag a microbe or an infected cell for attack by other parts of the immune system. The binding of an antibody can also neutralize its antigen target directly by, for example, blocking a part of a microbe that is essential for its survival and growth in the body. The production of these antibodies is the main function of the humoral immune system.[2]
Each antibody binds to a specific antigen; an interaction similar to a lock and key.
More specifically, antibodies are glycoproteins belonging to the immunoglobulin superfamily; the terms antibody and immunoglobulin are often used interchangeably.[3] They can occur in two forms, a soluble form that is secreted from cells, and a membrane-bound form that is attached to the surface of a B cell and is called the B cell receptor (BCR). The BCR is only found on the surface of B cells and binding of antigen to these proteins allows the B cells to detect when the antigen is present in the body. Once the B cells bind to their antigens and are activated, they differentiate into either antibody factories called plasma cells, or memory cells that will survive in the body for years afterwards, allowing the organism to remember that antigen and respond faster upon future exposures.[4] In most cases, interaction of the B cell with a T helper cell is necessary to produce full activation of the B cell and, therefore, antibody generation following antigen binding.[5] Soluble antibodies are released into the blood and tissue fluids, as well as many secretions to continue to survey for invading microorganisms.
Contents
1. Immunoglobulin isotypes
2. Structure of the antibody
2.1. Heavy chain
2.1.1. In mammals
2.1.2. In fish
2.2. Light chain
2.3. Fc region
2.4. Fab region
3. Immunoglobulin diversity
3.1. Peptide bond formation
3.2. Somatic hypermutation
3.3. Class switching
4. Affinity versus avidity
5. Function
5.1. Neutralization
5.2. Agglutination
5.3. Activation of complement
5.4. Activation of effector cells
6. Medical applications
6.1. RHOGAM antibodies
7. Research applications
8. References
1. Immunoglobulin isotypes
1.1. Functions in proteins
In mammals there are five types of antibody: IgA, IgD, IgE, IgG, and IgM (where Ig stands for immunoglobulin). In addition, there are 4 IgG and 2 IgA subtypes present in humans.[1] These are classified according to differences in their heavy chain constant domains.[3] Each immunoglobulin class differs in its biological properties and locations, and has evolved to deal with different antigens.[6]
• IgA can be found in areas containing mucus (e.g. in the gut, in the respiratory tract or in the urogenital tract) and prevents the colonization of mucosal areas by pathogens.[7]
• IgD functions mainly as an antigen receptor on B cells.[8]
• IgE binds to allergens and triggers histamine release from mast cells (the underlying mechanism of allergy) and also provides protection against parasitic worms.[2]
• IgG (in its four forms) provides the majority of antibody-based immunity against invading pathogens.[2]
• IgM is expressed on the surface of B cells and also in a secreted form with very high affinity for eliminating pathogens in the early stages of B cell mediated immunity (i.e. before there is sufficient IgG to do the job).[2][8]
Immature B cells express only IgM on their cell surface (this is the surface bound form, not the secreted form of immunoglobulin). Once the naive B cell reaches maturity, it can express both IgM and IgD on its surface – it is the co-expression of both these immunoglobulin isotypes that renders the B cell ‘mature’ and ready to respond to antigen.[9] Following engagement of the immunoglobulin molecule with an antigen, the B cell activates, and begins to divide and differentiate into an antibody producing cell (sometimes called a plasma cell). In this activated form, the B cell produces immunoglobulin in a secreted form rather than a membrane-bound form. Some of the daughter cells of the activated B cells undergo isotype switching, a mechanism by which the B cell begins to express the other Ig heavy chains and thus produce the IgD, IgA or (more commonly) IgG antibody isotypes.[10]
2. Structure of the antibody
Immunoglobulins are heavy plasma proteins, often with sugar chains added to amino acid residues by N-linked glycosylation (all antibodies) and occasionally O-linked glycosylation (e.g. IgA1 and IgD).[11] In other words, they are glycoproteins. The basic unit of each antibody is a monomer (one Ig unit) but the secreted antibody can also be dimeric (with two Ig units as with IgA), tetrameric (with four Ig units, like teleost fish IgM), or pentameric (with five Ig units, like mammalian IgM).[12] The monomer is a “Y”-shaped molecule that consists of four polypeptide chains; two identical heavy chains and two identical light chains connected by disulfide bonds.[6] The tip of the Y is important for binding to antigen, while the base of the Y is important for binding to specific receptors (such as Fc receptors) to allow activation immune responses appropriate for a given antigen.[13]
2.1. Heavy chain
2.1.1. In mammals
There are five types of mammalian immunoglobulin heavy chain: γ, δ, α, μ and ε.[1] They define classes of immunoglobulins. Heavy chains α and γ have approximately 450 amino acids, while μ and ε have approximately 550 amino acids.[1]
Each heavy chain has two regions:
• a constant region (which is the same for all immunoglobulins of the same class but differs between each class of immunoglobulins).
• Heavy chains γ, α and δ have a constant region composed of three tandem (in a line next to each other) immunoglobulin domains but also have a hinge region for added flexibility.[6]
• Heavy chains μ and ε have a constant region composed of four immunoglobulin domains.[1]
• a variable region that differs between different B cells, but is the same for all immunoglobulins produced by the same B cell or B cell clone. The variable domain of any heavy chain is composed of a single immunoglobulin domain. These domains are about 110 amino acids long.
2.1.2. In fish
Jawed fish appear to be the most primitive animals that are able to make antibodies like those described for mammals.[14] However, fish do not have the same repertoire of antibodies that mammals possess.[15] Three distinct Ig heavy chains have so far been identified in bony fish.
• The first identified was the μ (or mu) heavy chain that is present in all jawed fish and is the heavy chain for what is thought to be the primordial immunoglobulin. The resulting antibody, IgM, is secreted as a tetramer (containing four polypeptide chains) in teleost fish instead of the typical pentamer (containing five polypeptide chains) found in mammals and sharks.
• The heavy chain (δ) for IgD was identified initially from the channel catfish and Atlantic salmon and is now well documented for many teleost fish.[16]
• The third teleost Ig heavy chain gene was identified very recently and does not resemble any of the heavy chains so far described for mammals. This heavy chain, identified in both rainbow trout (τ)[17] and zebrafish (ζ),[18] could potentially form a distinct antibody isotype (IgT or IgZ) that may precede IgM in evolutionary terms.
Similar to the situation observed for bony fish, three distinct Ig heavy chain isotypes have been identified in cartilaginous fish. With the exception of IgM, these Ig heavy chain isotypes appear to be unique to cartilaginous fish and are designated IgM, IgW (also called IgX or IgNARC) and IgNAR.[19]
2.2. Light chain
There are only two types of light chain in mammals, λ and κ.[1] Other types of light chains are found in lower vertebrates as the Ig-Light-Iota chain in Chondrichthyes and Teleostei.[20] In each antibody, only one type is present and the two chains are identical. Each light chain has two successive domains: one constant and one variable domain. The approximate length of a light chain is from 211 to 217 amino acids.[1]
Camelids are unique among all other mammals in that they have fully functional immunoglobulins which consist of two heavy chains, but lacking the light chains usually paired with each heavy chain.[21] Although camels also have classical four-chain IgG1 antibodies, a splicing defect in the constant IGHG2 and IGHG3 genes leads to the absence of the CH1 domain in the heavy chain.[22] The functional role of this separate repertoire is unknown as yet. Apart from providing insight into immunoglobulin structure and antigen recognition in the absence of light chain CDR’s, these unusual antibodies can also be exploited to generate antibody fragments even smaller than single chain variable fragments, but much more stable.[23]
2.3. Fc region
The Fc region (Fragment, crystallizable), is derived from the stem of the “Y,” and is composed of two heavy chains that each contribute two to three constant domains (depending on the class of the antibody).[1] Fc binds to various cell receptors and complement proteins. In this way, it mediates different physiological effects of antibodies (opsonization, cell lysis, degranulation of mast cells, basophils and eosinophils and other processes).[6][24]
2.4. Fab region
Each end of the forked portion of the “Y” on the antibody is called the Fab region (Fragment, antigen binding). It is composed of one constant and one variable domain of each of the heavy and the light chain.[25] These domains shape the paratope—the antigen binding site—at the amino terminal end of the monomer. The two variable domains bind the epitope on their specific antigens.
In an experimental setting, Fc and Fab fragments can be generated in the laboratory. The enzyme papain can be used to cleave an immunoglobulin monomer into two Fab fragments and an Fc fragment. The enzyme pepsin cleaves below hinge region, so a F(ab’)2 fragment and a Fc fragment is formed. The variable regions of the heavy and light chains can be fused together to form a single chain variable fragment (scFv), which is only half the size of the Fab fragment yet retains the original specificity of the parent immunoglobulin.[26]
3. Immunoglobulin diversity
The success of antibodies to recognize and eradicate many different types of microbe requires their wide diversity; antibodies vary from one another in their amino acid composition allowing them to interact with different antigens.[27] An individual vertebrate possesses a large number of different antibodies, each capable of binding to a distinct antigen or epitope from a foreign object. However, although a huge number of different antibodies are generated, there is not an equally large array of genes available, in a single individual, to make such a huge repertoire of antibodies. Vertebrate B cells have several mechanisms that allow them to generate large antibody diversity from a relatively small number of immunoglobulin genes.[28]
3.1. V(D)J recombination
Somatic recombination, also known as V(D)J recombination, of immunoglobulins involves the random selection and combination of genes encoding each segment of the immunoglobulin variable region in a manner that generates a huge repertoire of antibodies with different paratopes. These segments are called variable (V), diversity (D) and joining (J) segments.[28] V, D and J segments are found in Ig heavy chains but only V and J segments are found in Ig light chains. Multiple copies of the V, D and J segments exist tandemly arranged in the genomes of mammals. Their selection for recombination within the individual B cell is also called gene rearrangement.[29] A B cell that successfully produces a functional immunoglobulin gene during its V(D)J recombination will suppress the expression of any other variable region gene by a process known as allelic exclusion.[30] Thus, the variable regions of all the immunoglobulin molecules within one given B cell will be the same, although the constant domains of the heavy chains can differ.[1] The diversity generated by this mechanism in the variable region of the heavy chain – to be specific, in the area that these V, D and J genes encode, otherwise known as the complementarity determining region 3 (CDR3) – provides the vertebrate immune system its ability to bind so many distinct antigens.
3.2. Somatic hypermutation
A further mechanism for generating antibody diversity exists for the mature B cell after antigen stimulation. Activated B cells are more prone to somatic hypermutation in their immunoglobulin variable chain genes.[31] This generates slight changes in the amino acid sequence of the variable domains of both the light and heavy chains between clones of the same activated B cell, and ultimately, differences in the affinity or strength of interaction that the B cell has with its specific antigen.[32] Thus, B cells expressing immunoglobulins with higher affinity for the antigen will outcompete those with weaker immunoglobulin for function and survival in a process known as affinity maturation.[33]
3.3. Class switching
Isotype switching (or class switching) occurs after the process of V(D)J recombination and following activation of the mature B cell to generate the different classes of antibody, all with the same variable domains as the original immunoglobulin generated in the immature B cell during recombination, but possessing distinct constant domains in their heavy chains.[29]
4. Affinity versus avidity
Depending on the structure of the antibody (which varies with the isotype) and that of the antigen, an antibody may have only one binding interaction with the antigen (monovalent) or multiple simultaneous interactions (multivalent).[1]
Affinity is the binding strength of a single antibody – antigen interaction.
Avidity is the compound affinity of multiple antibody – antigen interactions when more than one takes place between the two molecules. That is, avidity is the apparent affinity of the antigen – antibody binding in these cases, not the true affinity.
Avidity can be orders of magnitude greater than affinity, helping for instance poorly affinity-matured but highly multivalent IgM still bind antigen efficiently.[34]
5. Function
Since antibodies exist freely in the bloodstream or bound to cell membranes, they are said to be part of the humoral immune system. The circulating antibodies are produced by clonal B cells that are specific to only one antigen (e.g., a virus hull protein fragment). In binding their specific antigens, the antibodies can cause agglutination and precipitation of antibody-antigen products primed for phagocytosis by macrophages and other cells, block viral receptors, and stimulate other immune responses, such as the complement pathway.[35]
5.1. Neutralization
Antibodies that recognize viruses can block these by binding them directly.[2] In doing so, the virus will be unable to dock to its preferred receptor on a host cell in order to infect it. Some antibodies, like IgA, also directly bind to microbes in mucus to prevent the colonization of mucosal tissues. Some antibodies, like those in antivenoms, neutralize toxins by binding to them.[36] Problems can occur with certain viruses if antibody neutralization is inadequate. For example, when certain viruses such as the HIV, are not completely covered by neurtralizing antibody, the antibodies can enhance viral infectivity instead of inhibiting it; HIV prefers to infect the cells that bind to antibodies.[37] Antibodies cannot attack pathogens within cells, and certain viruses (such as HIV, HSV and HBV) “hide” inside cells for long periods of time to avoid them.[2] This is the reason for the chronic nature of many minor skin diseases (such as cold sores); any given outbreak is quickly suppressed by the immune system, but the infection is never truly eradicated because some cells retain viruses that will resume the symptoms later.[2]
5.2. Agglutination
Antibodies are clonally generated for binding single specific antigens, which may compose specific molecules on the surfaces of viruses or cells. The antibodies can link these viruses or cells together, causing them to agglutinate (coagulate) so phagocytes can capture them.[2]
5.3. Activation of complement
Antibodies that bind to surface antigens on, for example a bacterium, will bind the first component of the complement system with their Fc region and initiate activation of the “classical” complement system.[35] This results in the killing of bacteria in two ways.[2] First, the binding of the antibody and complement molecules marks the microbe for ingestion by phagocytes in a process called opsonization. These phagocytes are attracted by some of the complement molecules that are generated in the complement cascade. Secondly, some complement system components form a membrane attack complex to assist the antibodies to kill bacteria directly.[38]
5.4. Activation of effector cells
Some cells (e.g. Mast cells and phagocytes) have specific receptors on their cell surface for binding antibodies. These are called Fc receptors, and, as the name suggests, these receptors interact with the Fc region of some antibodies (e.g. IgA, IgG, IgE). The engagement of a particular antibody with the Fc receptor on a particular cell will trigger the effector function of that cell (e.g. phagocytes will phagocytose, mast cells will degranulate) that will ultimately result in destruction of the invading microbe. The Fc receptors are isotype-specific, which gives a great flexibility to the immune system, because different situations require only certain immune mechanisms to respond to antigens.[1]
6. Medical applications
Detection of particular antibodies is a very common form of medical diagnostics. Serology depends on these methods.[39] Autoimmune disorders can often be traced to antibodies that bind the body’s own epitopes; many can be detected through blood tests. Antibodies directed against RBC surface antigens in immune mediated hemolytic anemia can be detected with the Coombs test.[40] The Coombs test is also used for antibody screening in blood transfusion preparation and also for antibody screening in antenatal women.[40]
“Targeted” monoclonal antibody therapy is already being employed to treat diseases such as rheumatoid arthritis,[41] multiple sclerosis,[42] psoriasis,[43] and in many forms of cancer including non-Hodgkin’s lymphoma,[44] colorectal cancer, head and neck cancer and breast cancer.[45]
Some immune deficiencies, such as X-linked agammaglobulinemia and hypogammaglobulinemia result in partial or complete lack of antibodies.[46] These diseases are often treated by inducing a short term form of immunity called passive immunity. Passive immunity is achieved through the transfer of ready made antibodies in the form of human or animal serum, pooled immunoglobulin or monoclonal antibodies, into the affected individual.[47]
Elevations in the different classes of immunoglobulins are sometimes useful in determining the cause of liver damage in patients whom the diagnosis is unclear.[3] For example, elevated IgA indicates alcoholic cirrhosis, elevated IgM indicates viral hepatitis and primary biliary cirrhosis, while IgG is elevated in viral hepatitis, autoimmune hepatitis and cirrhosis.
6.1. RHOGAM antibodies
RHOGAM antibodies are a trade name for Rho(D) Immune Globulin antibodies specific to the human Rh D antigen.[48] They normally administered as part of a pre-natal treatment regimen to prevent any sensitization that may occur when a Rhesus-negative mother has a fetus that is Rhesus-positive. The Rhesus factor (a.k.a. D antigen) is an antigen found on red blood cells in the blood. It is the second most significant risk issue in a blood transfusion, next to the ABO blood type. People that are Rh+ have this antigen on their red blood cells. People that are Rh- don’t have this antigen on their red blood cells.
In the course of regular childbirth delivery trauma or other prenatal complications, blood from the fetus occasionally enters the mother’s system. In the case of an Rh-incompatible mother and child, blood mixing from this trauma may ‘sensitize’ the Rh-negative mother to the Rh antigen, putting the remainder of the pregnancy, and any subsequent pregnancies, at risk for hemolytic disease of the newborn.[49]
In contrast, treatment of an unsensitized mother with RhoGAM antibodies prior to and immediately after trauma and delivery will immediately destroy any Rh antigen incidentally in the mother’s system from the fetus. Importantly, this will happen before the antigen can stimulate the mother’s memory-mediated immune response B cells to “remember” Rh antigen. Therefore, her humoral immune system will never be stimulated to make anti-Rh antibodies, and will not attack the current, or any potential subsequent, baby’s Rhesus antigens. RhoGAM prevents ‘sensitization’ that can lead to Rh disease, but does not prevent or treat the underlying disease itself.
7. Research applications
In research, antibodies are used in many applications. The most common is the identification and localization of intracellular and extracellular proteins. Antibodies may be used to differentiate cell types,[50] to separate proteins (and anything bound to them) from the other molecules in a cell lysate,[51] to identify proteins after electrophoresis,[52] or to examine protein expression in tissues.[50]
These purified antibodies are often produced by injecting the antigen into a small mammal, such as a mouse or rabbit. Sometimes, in order to obtain large quantity of antibodies, goats, sheep, or horses are used. Blood isolated from these animals contains polyclonal antibodies — multiple antibodies that bind to the same antigen. The serum, also known as the antiserum, because it now contains the desired antibodies, is commonly purified with Protein A/G purification or antigen affinity chromatography.[53] If the lymphocytes that produce the antibodies can be isolated and immortalized, then a monoclonal antibody can be obtained.
In biochemical assays for disease diagnosis,[54] a titer of Epstein-Barr virus or Lyme disease will look for antibodies produced by the body that are specific to those antigens in the blood. If those antibodies are not present, either the person has not been infected, or the infection occurred a very long time ago, and the antibodies have naturally decayed.
8. References
1. Janeway CA, Jr. et al (2001). Immunobiology., 5th ed., Garland Publishing.
2. Pier GB, Lyczak JB, Wetzler LM (2004). Immunology, Infection, and Immunity. ASM Press.
3. Rhoades RA, Pflanzer RG (2002). Human Physiology, 4th ed., Thomson Learning.
4. Borghesi L, Milcarek C (2006). “From B cell to plasma cell: regulation of V(D)J recombination and antibody secretion”. Immunol Res 36 (1-3): 27-32.
5. Parker D (1993). “T cell-dependent B cell activation”. Annu Rev Immunol 11: 331-60.
6. Woof J, Burton D (2004). “Human antibody-Fc receptor interactions illuminated by crystal structures”. Nat Rev Immunol 4 (2): 89-99.
7. Underdown B, Schiff J (1986). “Immunoglobulin A: strategic defense initiative at the mucosal surface”. Annu Rev Immunol 4: 389-417.
8. Geisberger R, Lamers M, Achatz G (2006). “The riddle of the dual expression of IgM and IgD”. Immunology 118 (4): 429-37.
9. Goding J. “Allotypes of IgM and IgD receptors in the mouse: a probe for lymphocyte differentiation”. Contemp Top Immunobiol 8: 203-43.
10. Neuberger M, Ehrenstein M, Rada C, Sale J, Batista F, Williams G, Milstein C (2000). “Memory in the B-cell compartment: antibody affinity maturation”. Philos Trans R Soc Lond B Biol Sci 355 (1395): 357-60.
11. Mattu T, Pleass R, Willis A, Kilian M, Wormald M, Lellouch A, Rudd P, Woof J, Dwek R (1998). “The glycosylation and structure of human serum IgA1, Fab, and Fc regions and the role of N-glycosylation on Fc alpha receptor interactions”. J Biol Chem 273 (4): 2260-72.
12. Roux K (1999). “Immunoglobulin structure and function as revealed by electron microscopy”. Int Arch Allergy Immunol 120 (2): 85-99.
13. ^ Huber R (1980). “Spatial structure of immunoglobulin molecules”. Klin Wochenschr 58 (22): 1217-31. PMID 6780722.
14. Fish heavy chain and light chain genes
15. Eva Bengtén, L. William Clem, Norman W. Miller, Gregory W. Warr and Melanie Wilson. Channel catfish immunoglobulins: Repertoire and expression. Developmental & Comparative Immunology, Volume 30, Issues 1-2, Antibody repertoire development, 2006, Pages 77-92.
16. Stein Tore Solem and Jørgen Stenvik. Antibody repertoire development in teleosts–a review with emphasis on salmonids and Gadus morhua L. Developmental & Comparative Immunology, Volume 30, Issues 1-2, Antibody repertoire development, 2006, Pages 57-76.
17. J.D. Hansen, E.D. Landis and R.B. Phillips. Discovery of a unique Ig heavy-chain isotype (IgT) in rainbow trout: Implications for a distinctive B cell developmental pathway in teleost fish. Proceedings of the National Academy of Sciences U S A. Volume 102, Issue 19, 2005, pages 6919-24.
18. N. Danilova, J. Bussmann, K. Jekosch, L.A Steiner. The immunoglobulin heavy-chain locus in zebrafish: identification and expression of a previously unknown isotype, immunoglobulin Z. Nature Immunology, Volume 6, Issue 3, 2005, pages 295-302.
19. H. Dooley and M.F. Flajnik. Antibody repertoire development in cartilaginous fish. Developmental & Comparative Immunology, Volume 30, Issues 1-2, Antibody repertoire development, 2006, Pages 43-56.
20. IMGT Index Antibodies (or Immunoglobulins).
21. Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa E, Bendahman N, Hamers R (1993). “Naturally occurring antibodies devoid of light chains”. Nature 363 (6428): 446-8.
22. Conrath K, Wernery U, Muyldermans S, Nguyen V (2003). “Emergence and evolution of functional heavy-chain antibodies in Camelidae”. Dev Comp Immunol 27 (2): 87-103.
23. Muyldermans S (2001). “Single domain camel antibodies: current status”. J Biotechnol 74 (4): 277-302.
24. Heyman B (1996). “Complement and Fc-receptors in regulation of the antibody response”. Immunol Lett 54 (2-3): 195-9.
25. Putnam FW, Liu YS, Low TL (1979). “Primary structure of a human IgA1 immunoglobulin. IV. Streptococcal IgA1 protease, digestion, Fab and Fc fragments, and the complete amino acid sequence of the alpha 1 heavy chain”. J Biol Chem 254 (8): 2865-74.
26. Wu A, Yazaki P (2000). “Designer genes: recombinant antibody fragments for biological imaging”. Q J Nucl Med 44 (3): 268-83.
27. Mian I, Bradwell A, Olson A (1991). “Structure, function and properties of antibody binding sites”. J Mol Biol 217 (1): 133-51.
28. Nemazee D (2006). “Receptor editing in lymphocyte development and central tolerance”. Nat Rev Immunol 6 (10): 728-40.
29. Eleonora Market, F. Nina Papavasiliou (2003) V(D)J Recombination and the Evolution of the Adaptive Immune System PLoS Biology1(1): e16.
30. Bergman Y, Cedar H (2004). “A stepwise epigenetic process controls immunoglobulin allelic exclusion”. Nat Rev Immunol 4 (10): 753-61.
31. Diaz M, Casali P (2002). “Somatic immunoglobulin hypermutation”. Curr Opin Immunol 14 (2): 235-40.
32. Honjo T, Habu S (1985). “Origin of immune diversity: genetic variation and selection”. Annu Rev Biochem 54: 803-30.
33. Neuberger M, Ehrenstein M, Rada C, Sale J, Batista F, Williams G, Milstein C (2000). “Memory in the B-cell compartment: antibody affinity maturation”. Philos Trans R Soc Lond B Biol Sci 355 (1395): 357-60.
34. Pancook J, Beuerlein G, Pecht G, Tang Y, Nie Y, Wu H, Huse W, Watkins J (2001). “In vitro affinity maturation of human IgM antibodies reactive with tumor-associated antigens”. Hybrid Hybridomics 20 (5-6): 383-96.
35. Ravetch J, Bolland S (2001). “IgG Fc receptors”. Annu Rev Immunol 19: 275-90.
36. Chippaux J, Goyffon M (1998). “Venoms, antivenoms and immunotherapy”. Toxicon 36 (6): 823-46.
37. Weiss R (2001). “Gulliver’s travels in HIVland”. Nature 410 (6831): 963-7.
38. Rus H, Cudrici C, Niculescu F (2005). “The role of the complement system in innate immunity”. Immunol Res 33 (2): 103-12.
39. Animated depictions of how antibodies are used in ELISA assays
40. Dean, Laura (2005). “Chapter 4: Hemolytic disease of the newborn”, Blood Groups and Red Cell Antigens. NCBI Bethesda (MD): National Library of Medicine (US),.
41. Feldmann M, Maini R (2001). “Anti-TNF alpha therapy of rheumatoid arthritis: what have we learned?”. Annu Rev Immunol 19: 163-96.
42. Doggrell S (2003). “Is natalizumab a breakthrough in the treatment of multiple sclerosis?”. Expert Opin Pharmacother 4 (6): 999-1001.
43. Krueger G, Langley R, Leonardi C, Yeilding N, Guzzo C, Wang Y, Dooley L, Lebwohl M (2007). “A human interleukin-12/23 monoclonal antibody for the treatment of psoriasis”. N Engl J Med 356 (6): 580-92.
44. Plosker G, Figgitt D (2003). “Rituximab: a review of its use in non-Hodgkin’s lymphoma and chronic lymphocytic leukaemia”. Drugs 63 (8): 803-43.
45. Vogel C, Cobleigh M, Tripathy D, Gutheil J, Harris L, Fehrenbacher L, Slamon D, Murphy M, Novotny W, Burchmore M, Shak S, Stewart S (2001). “First-line Herceptin monotherapy in metastatic breast cancer”. Oncology 61 Suppl 2: 37-42.
46. LeBien T (2000). “Fates of human B-cell precursors”. Blood 96 (1): 9-23.
47. Microbiology and Immunology On-Line Textbook: USC School of Medicine
48. Fung Kee Fung K, Eason E, Crane J, Armson A, De La Ronde S, Farine D, Keenan-Lindsay L, Leduc L, Reid G, Aerde J, Wilson R, Davies G, Désilets V, Summers A, Wyatt P, Young D (2003). “Prevention of Rh alloimmunization”. J Obstet Gynaecol Can 25 (9): 765-73.
49. Urbaniak S, Greiss M (2000). “RhD haemolytic disease of the fetus and the newborn”. Blood Rev 14 (1): 44-61.
50. Brehm-Stecher B, Johnson E (2004). “Single-cell microbiology: tools, technologies, and applications”. Microbiol Mol Biol Rev 68 (3): 538-59.
51. Williams N (2000). “Immunoprecipitation procedures”. Methods Cell Biol 62: 449-53.
52. Kurien B, Scofield R (2006). “Western blotting”. Methods 38 (4): 283-93.
53. Kabir S. “Immunoglobulin purification by affinity chromatography using protein A mimetic ligands prepared by combinatorial chemical synthesis”. Immunol Invest 31 (3-4): 263-78.
54. Animated depictions of how antibodies are used in ELISPOT assays


